16 Metabolic Activities B: Physiological Characteristics Results & Interpretation
Now that your bacterial species has had the time to grow and metabolize on several types of media, it is time to collect the results of the biochemical assays to determine physiological characteristics. When performing the Unknown procedure, you will be able to interpret all data, and identify the bacterium.
Some biochemical test results are simply interpretation of appearance due to the medium having a pH indicator. For other tests, chemical reagents must be added to the inoculated medium so that the results can be seen. The procedure of this exercise will go through the steps that need to be done in order to receive results of the biochemical assays for interpretation along with an explanation of the actual metabolism that is being carried out by the bacteria. Photos of positive and negative results are available.
MATERIALS
Per pair of students for “Knowns”
Blue rack and empty 20 mm test tube
Coffee can and wax pencil
Green pi-pump and 5 ml serological pipette
Your inoculated tubes and plates from previous lab:
| TSA agar | (agar plate, Trypticase Soy) | Nitrate broth | (yellow cap broth tube) |
| Milk agar | (agar plate, opaque white) | MR-VP broth | (black cap broth tube) |
| Starch agar | (agar plate, black line on lid) | PR Sucrose | (blue cap broth tube) |
| Simmon’s Citrate agar | (screw-top agar tube) | PR Dextrose | (green cap broth tube) |
| SIM medium agar | (clear cap agar tube) | PR Mannitol | (red cap broth tube) |
| Urea broth | (white cap broth tube) | PR Lactose | (yellow cap broth tube) |
This first stage is the “Knowns” run-through the tests will show how to interpret the tests. Students will do this work in pairs.
The second stage is the “Unknowns” where the actual identification of an unknown culture occurs. In that stage, each student will perform these procedures, independently.
PROCEDURES FOR METABOLIC ACTIVITY TESTS:
- Collect your test tubes and plates from the incubator. Remove your test tubes from the coffee can, and place them in a blue rack.
- Two tests take longer to produce accurate results (Voges-Proskauer and Oxidase). Start them early, to save time. These two tests each use a medium that also covers another test. Start with:
a. MR-VP broth (Voges-Proskauer test and Methyl Red test)
b. TSA plate (Oxidase test and Catalase test)
MR-VP Broth (black cap). Used for two tests:
- Using a 5 ml serological pipette and a green pi-pump, remove 2 ml from the MR-VP tube, and transfer aseptically into an empty 20 mm test tube.
- Label the 2 ml tube VP with the wax pencil.
Voges-Proskauer test
- Wear gloves. Add 10 drops of each: Barritt’s reagents A & B into the 2 ml.
- Shake tube left to right to oxygenate bacteria for 1 to 2 minutes.
- Set aside for at least 30 minutes. Do not read immediately. Then observe.
- Label the remaining 8 ml in the black cap tube MR.
Methyl Red test
- Add ¼ dropper full of methyl red reagent and shake. Then observe color.
MR-VP broth contains a large quantity of glucose. Some bacteria are able to ferment glucose through different pathways, such as butylene glycol pathway and mixed acid pathway. The butylene glycol pathway is where acid products (pyruvate) are quickly converted to the alcohols acetoin and 2,3-butanediol. This is what the Voges-Proskauer test is testing for by adding Barritt’s A and B. The reagents oxidize the acetoin, if present, producing a red to burgundy color for a positive result. (Figure 16.1).
The mixed acid pathway is when glucose is fermented to produce several acids, such as lactic, acetic, and formic acids, that are fairly stable, lowering the pH of the medium. Methyl red pH indicator is added to the medium. The change to a red color, showing a low pH of at least 4.4, is a positive result. (Figure 16.2).
|
 |
 |
| Figure 16.1: Voges Proskauer test: The tube on the left is positive (red/ burgundy color) while the tube on the right is negative. | Figure 16.2: Methyl red test: The tube on the left is positive (red color) while the tubes on the right is negative. This test is based on pH. Yellow and orange are not acidic. |
TSA Plate: Used for two tests
Oxidase test
This test identifies bacteria that produce the enzyme cytochrome oxidase.
Cytochrome oxidase participates in the electron transport chain of aerobic respiration, transferring electrons to oxygen and producing water. In aerobic respiration, oxygen is the terminal electron acceptor. This enzyme is detected by an oxy-swab which contains the oxidase reagent of an artificial electron acceptor. A positive for cytochrome oxidase is a purple color. (Figure 16.3).
- Add a drop of distilled water directly onto an oxy-swab.
- Using one streak on the TSA plate, roll the oxy-swab along the outside edge of bacterial streak.
- Press swab against inside of petri plate lid to saturate bacteria with reagent and get past the glycocalyx.
- Wait at least 1 minute before looking at result.
Catalase test
The catalase test is used to differentiate bacteria that are aerobes or facultative anaerobes from bacteria that are obligate anaerobes, microaerophiles or aerotolerant anaerobes. During aerobic respiration, oxygen is used as a terminal electron acceptor, forming water. However, hydrogen peroxide is a byproduct, which is a reactive oxygen species which can react with parts of the cell and cause damage. Aerobes and facultative anaerobes can use enzymes to detoxify oxygen. For example, the enzyme catalase breaks down hydrogen peroxide into water and harmless oxygen gas. To detect for the enzyme catalase, apply hydrogen peroxide to a bacterial streak. A positive result is bubbles because this is the oxygen being released from the bacteria. (Figure 16.4).
- Add a dropper full of hydrogen peroxide directly to the other streak on the TSA plate (not used for the oxidase test) and observe.
 |
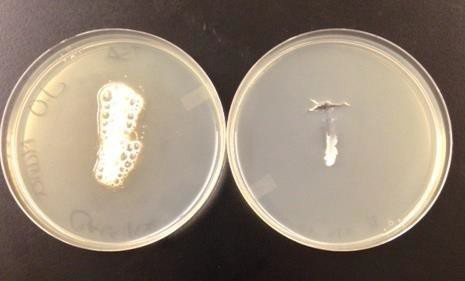 |
| Figure 16.3: Oxidase test: The swab on the left is positive (dark purple) while the swab on the right is negative (no color change). | Figure 16.4: Catalase test: The plate on the left is positive (bubbles) while the plate on the right is negative (no bubbles). |
Starch Hydrolysis
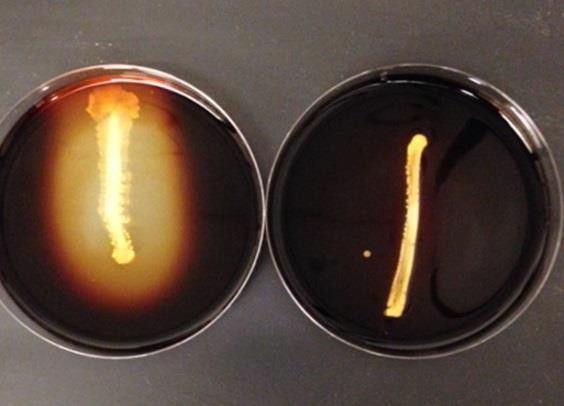
The differential medium starch agar (black line on lid) is used to distinguish bacteria that can hydrolyze, or break down, starch into mono and disaccharides from bacteria that cannot. Bacteria produce the exoenzyme amylase, which breaks the bonds apart in the starch. Iodine must be added to the plate to detect that the reaction occurred. Iodine complexes with starch to give a black or dark purple color, but if the starch has been broken down into simple sugars, a yellow to brown color will be present. (Figure 16.5). Amylase is an exoenzyme, so the cases where the culture itself is blocking the iodine from entering the agar do not count as a sign of amylase activity.
- Add iodine directly to the surface of the plate. Completely cover the plate.
- Observe color change.
 |
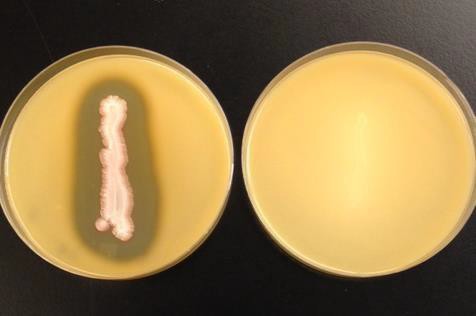 |
| Figure 16.5: Starch hydrolysis: The plate on the left is positive (clearing) while the plate on the right is negative (no clearing). | Figure 16.6: Casein hydrolysis: The plate on the left is positive (clearing) while the plate on the right is negative (no clearing) |
Casein Hydrolysis
The differential medium milk agar is used to distinguish bacteria that can hydrolyze the protein casein into amino acids from bacteria that cannot. Casein is the major protein in milk, giving it its white color. Bacteria hydrolyze casein by producing the exoenzyme caseinase, which breaks down the peptide bonds between the amino acids. If casein is broken down in the milk agar, the opaque white color of the milk now appears as a clearing due to being amino acids and no longer a polypeptide (protein). (Figure 16.6).
- Observe plate for clearing.
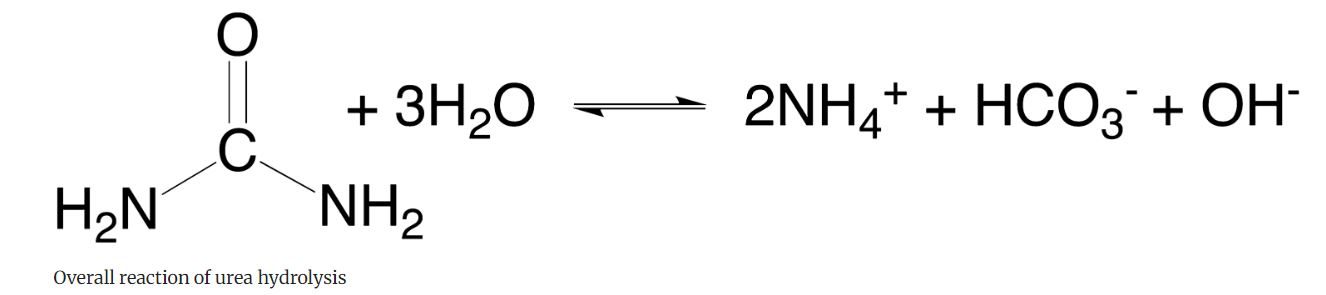
Urea Hydrolysis
Urea is a waste product of animal metabolism that is broken down by a number of bacteria. Urea broth is a differential medium that contains a pH indicator that tests the ability of bacteria to produce the exoenzyme urease that hydrolyzes urea into ammonia and carbon dioxide. This enzyme is produced by some bacteria.
Ammonia takes up H+ to become NH4+ and increases the pH so in effect the phenol red pH indicator changes from yellow to a bright pink as a positive result. (Figure 16.7).
- Observe color of medium.
  |
 |
| Figure 16.7: Urea Hydrolysis: The tube on the left is positive (hot pink color) while the tube on the right is negative (no color change/ orange). | Figure 16.8: Nitrate Reduction: Tube A is positive (red color). Tube B and C both have Zinc granules added. Tube B is positive (yellow color). Tube C is negative (red color). |
Nitrate Reduction
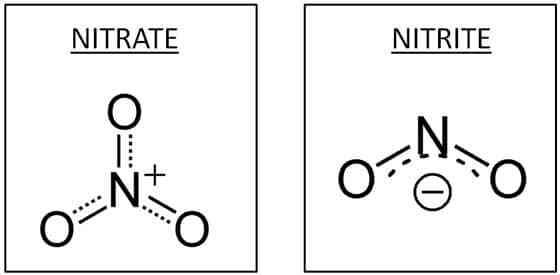
Some fermenters can reduce nitrate (NO3-) to nitrite (NO2-) in order to regenerating NAD+ from NADH. As fermenters, they get very little energy from their metabolism and die from their waste. These bacteria will leave behind a lot of NO2-. This is called dissimilatory nitrate reduction.
Some facultative anaerobes and obligate anaerobes use nitrate, not oxygen, as a terminal electron acceptor during anaerobic respiration. This type of anaerobic respiration is called denitrification. These bacteria have enzymes, including nitrate reductase, that transfer electrons from the electron transport chain to NO3-, partially reducing it to NO2- and then eventually reduce it to nitrous oxide (N2O) or nitrogen gas (N2). Neither NO3- nor NO2- remain, because of the much greater efficiency of respiration.
- Wearing gloves, Add 10 drops of each: Nitrate Reagents 1 & 2.
- Gently shake the tube from left to right.
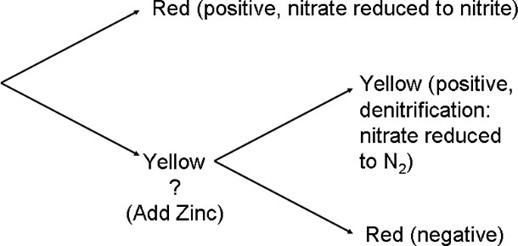
- If a red color is produced, then the bacterium is positive for nitrate reductase. (Figure 16.8).
- If a yellow color remains, another reagent has to be added to confirm a negative result.
- Add Zinc granules and shake. It may take time to see a red color. (You do not add Zinc granules to an already red positive tube.)
| Nitrate reagents 1 & 2 react with nitrite to make a red color. Zinc is a catalyst for reducing nitrate to nitrite, similar to dissimilatory nitrate reduction. Zinc checks for the original nitrate.
   |
|
Phenol Red Carbohydrate Broth
Phenol red carbohydrate broth is a differential medium used to detect for fermentation of specific carbohydrates, most commonly lactose, dextrose (glucose), mannitol, and sucrose. The ability of bacteria to ferment a specific sugar may be used to differentiate between genera and species. The medium contains the pH indicator phenol red, which turns yellow in the presence of acid (as low as pH 6.8), and a small inverted tube called the Durham tube to collect any gas that may be released during fermentation. (Figure 16.9)
The number of carbons in the acid produced determines gas production. For instance acetic acid production (2C) would be associated with gas production while lactic acid production (3C the same as pyruvate) would produce no gas.
- Observe color of media in the 4 tubes.
- Observe Durham tube, looking for gas in the Durham tube.


Figure 16.9: Phenol Red Carbohydrate Broth: For each set of three tubes, the tube on the left is positive for fermentation due to acid and gas (yellow and bubble in Durham tube) while the tube in the middle is only positive for acid (yellow). The tube on the right is negative for fermentation (red and no bubble). Color cap system: Yellow cap = lactose, Green cap = dextrose, Red cap = mannitol, Blue cap = sucrose.
h. SIM Medium. Used for 2 tests
SIM medium is a semisolid differential medium that is used for the detection of sulfur reduction, indole production, and motility. It is recommended for the differentiation for gram negative enteric bacilli (coliforms). We will be looking for the results of sulfur reduction and indole production only.
Sulfur Reduction
Coliforms degrade the protein casein that is in the SIM medium into amino acids. One particular amino acid is cysteine. SIM medium also contains an iron- containing compound and sulfur in the form of sodium thiosulfate. Sulfur reduction produces hydrogen sulfide gas in the anaerobic process. There are two different courses of action. These can be achieved depending on enzymes produced. The enzyme cysteine desulfurase breaks down cysteine to produce pyruvic acid, ammonia, and hydrogen sulfide gas. The other enzyme that serves in sulfur reduction is thiosulfate reductase. Sodium thiosulfate is a terminal electron acceptor in anaerobic respiration, and thiosulfate reductase catalyzes the reaction of reducing thiosulfate (sulfur) to the hydrogen sulfide gas product. The iron- containing compounding the SIM medium will react with the hydrogen sulfide gas to produce ferrous sulfide, showing a black precipitate to indicate a positive result. (Figure 16.10).
- Observe color of medium.
Indole test
SIM medium is also used to detect for the indole production. Bacteria partially breakdown the amino acid tryptophan to produce pyruvic acid, ammonia, and indole by using the enzyme tryptophanase. (Figure 16.11)
- Add ¼ to ½ of a dropper full of Kovac’s reagent to the tube in order to see a layer on top of the semisolid medium.
- Observe color of reagent.
 |
 |
| Figure 16.10: Hydrogen Sulfide Production: The tube on the left is positive (black precipitate) while the tube on the right is negative (no color change). | Figure 16.11: Indole Production: The tube on the left is positive (red color) while the tube on the right is negative
(no color change/ yellow). |
Citrate Utilization
Citrate utilization uses the medium Simmon’s Citrate agar that contains sodium citrate as a sole carbon source. It also contains inorganic ammonium salts as a sole nitrogen source. Citrate is metabolized in the citric acid cycle (or TCA) by the enzyme citrate permease. For uptake from the medium, citrate transporters must be able to handle the three negative charges (by symport or antiport) which will typically increase pH. The agar contains a pH indicator called bromthymol blue. In alkaline conditions, the green agar will turn to a deep blue color. (Figure 16.12).
- Observe color of medium.
 |
 |
| Figure 16.12: Citrate Utilization: The tube on the left is positive (deep blue color) while the tube on the right is negative (no color change/ green). Just a blue tinge at the top is also a negative. |
IMViC tests
Four of the metabolic tests can be grouped together naming them as the IMViC tests (I: indole test; M: methyl red test; V: Voges-Proskauer test; and C: citrate utilization test). These tests are used to differentiate bacteria that belong to the family Enterobacteriaceae for water analysis looking for fecal contamination, differentiating Escherichia coli and Klebsiella aerogenes. E. coli is found in human and animal intestines whereas K. aerogenes is a soil bacteria. These bacteria are morphologically and culturally similar. However, on these four tests, they physiologically differ. The two bacteria give exactly the opposite reactions.
| Indole | Methyl Red | Voges-Proskauer | Citrate | |
| E. coli | + | + | – | – |
| K. aerogenes | – | – | + | + |
Table 16.1: IMViC tests: Comparing the physiological difference between
E. coli and K. aerogenes.
Important:
After you have collected all your data, interpret the results and use the “Unknown Booklet of Organisms” to identify your unknown. Fill out your Descriptive Lab Chart, and discuss with your instructor your unknown identification. Do not discard any of your test tubes, plates, or oxy-swab until the unknown identification has been verified correctly. If you have your unknown identification incorrect, your instructor can analyze your tests to see if everything has been interpreted correctly.
Sometimes bacteria do not produce an enzyme that it typically should or vice versa. This may alter your deduction of the identification. Do not worry about a few mismatches, except for Gram-stain and cell shape. The metabolic tests can get you to a correct result even if a few of them don’t work correctly.
REFERENCES
Anaerobe Nitrate Reagent. (2017). Retrieved from Hardy Diagnostics: https://catalog.hardydiagnostics.com/cp_prod/Content/hugo/AnaerobeNitrateReagent.html
Brown, A. E. (2009). Benson’s Microbiological Applications: Laboratory Manual in General Microbiology. New York: McGraw Hill.
Chess, B. (2015). Laboratory Applications in Microbiology: A Case Study Approach. New York: McGraw Hill.
Leboffe, Michael. (2012). A Photographic Atlas for the Microbiology Laboratory (pp. 64-65). Englewood, CO: Morton Publishing Company.
Leboffe, Michael J. and Burton E. Pierce. (2011). A Photographic Atlas for the Microbiology Laboratory (pp. 93-94). Englewood, CO: Morton Publishing Company.
Mazzei, L., Musiani, F. & Ciurli, S. The structure-based reaction mechanism of urease, a nickel dependent enzyme: tale of a long debate. J Biol Inorg Chem 25, 829–845 (2020). https://doi.org/10.1007/s00775-020-01808-w
MR-VP tests. (n.d.). Retrieved from Microbugz: http://www.austincc.edu/microbugz/mrvp_test.php
Phenol Red w/ and w/o Carbohydrate w and w/o Durham tube. (2011, March 29). Retrieved from Thermo Fisher: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/IFU62202.pdf
SIM (Sulfide, Indole, Motility) Medium. (n.d.). Retrieved from Hardy Diagnostics: https://catalog.hardydiagnostics.com/cp_prod/Content/hugo/SIMMedium.htm

